Cort
Phoenix Rising Founder
- Messages
- 7,392
I admit I don't understand all the science. This is at the end of the Supplemental Sciences section - First they show that three CFS XMRV strains were very similar to the reference strain (VP62). They also show where the strains differ - which, would I think be important.
Maybe all the other strains after that were different - they were unlucky with those first strains..
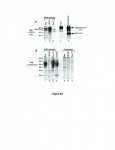
This next part is difficult but it appears to me to state that they could detect the VP 62 clone in their XMRV infected samples. They used lysates prepared using Raj(VP62), the LnCap cells they were using and another. If you'll look on the figure you'll see that XMRV shows up in all three of them..in fact in B it shows up better in the VP62 line than the LNCap line which could mean, I suppose, that VP62 does not show up well when you are using LNCap to grow the cells at least in some cases.
I don't know what all this means - and I grant that the VP62 clone is not the best - but it was in there, in some fashion, in the first study...Maybe you explain more about this.
Figure S1. Gag sequences of XMRV in CFS patients. Partial sequences (nt 6491017) from CFS XMRV strains WPI-1130, WPI-1138 and WPI-1169 in comparison to XMRV strains VP35, VP42 and VP62 derived from prostate cancer patients. The yellow highlighting denotes the differences from the reference strain (VP62).
Maybe all the other strains after that were different - they were unlucky with those first strains..
This next part is difficult but it appears to me to state that they could detect the VP 62 clone in their XMRV infected samples. They used lysates prepared using Raj(VP62), the LnCap cells they were using and another. If you'll look on the figure you'll see that XMRV shows up in all three of them..in fact in B it shows up better in the VP62 line than the LNCap line which could mean, I suppose, that VP62 does not show up well when you are using LNCap to grow the cells at least in some cases.
I don't know what all this means - and I grant that the VP62 clone is not the best - but it was in there, in some fashion, in the first study...Maybe you explain more about this.
Figure S3. Detection of cloned XMRV-VP62 using a rat mAb to SFFV Env and a goat antiserum to mouse NZB xenotropic MLV .
A. Lysates were prepared from XMRV-VP62-infected Raji (lane1), LNCaP (lane 2) or Sup-T1 (lane 3). Positive controls used were HCD-57 cells, a mouse erythroleukemia cell line expressing polytropic MLV gp70 Env (lane 4), and HCD-57 cells infected with SFFV, which also express SFFV gp55 Env (lane 5). WB analysis was carried out using rat anti-SFFV Env mAb 7C10. Molecular weight markers in kD are shown on the left.
B. Lysates were prepared from XMRV-VP62-infected Raji (lane 1), LNCaP (lane 2) or Sup-T1 (lane 3). Lysates from SFFV-infected mouse HCD-57 cells (lane 4) and from uninfected Raji, LNCaP and Sup-T1 are shown in lanes 5-7, respectively.