aquariusgirl
Senior Member
- Messages
- 1,736
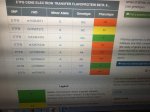 View attachment 26231 View attachment 26232 View attachment 26231 View attachment 26232
View attachment 26231 View attachment 26232 View attachment 26231 View attachment 26232I just installed a gene report on Livewello that seems to suggest I have some genetic weaknesses with the electron transport flavoprotein beta polypeptide gene.
A quick google reveals related conditions are (see below). Also, this gene has come up in autism research & I read that GAII has been associated with fatty infiltraition of the liver. And I have fatty liver.
Has this come up before?
Last edited:
