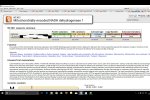
There are many different ways to sort the data. The first one is to see if you have any rare pathogenic mutations pop up in Enlis' already defined mutations.
After you've gone through the process to upload your data into the system, you'll see a screen like this. Next to the word genome, will be the names of the people in your system. Here, I've whited some family members names out.
If the above 1 genome was me, I'd click on the little button next to my name that looks like this:
Now it should look like this:
This is my version using my 23andme raw data. Whole exome data provides many times more variations than this. It's not unusual for a person to have this many.
I'd then click on the 627,912 in the above report to filter and sort those variations.
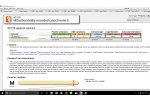
You should be on the Variation Filter Page. On the top right side, click on "Load Filter Set" and go down and click on "All Clinically Significant".
Then right under the tan box, there are buttons, at the top of the fifth column of buttons is "+ Allele Frequency". Click on that. I then slide it over 10% of the population having this variance allele. I use 10% as a starting point because sometimes fairly rare mutations (like 10%) combined with another rare mutation on the same gene can be the issue. Multiple issues on same gene can be the culprit. In addition, 10% in this sort is only going to leave you with a few dozen mutations that are pathogenic or a disease risk factor that you carry that are also rare.
Then push the little grey button on the left that says "Go".
This should bring up your variances. A word of caution here. Some of the issues will look like major issues, and then with some research into the mutation in the medical literature you'll find out the the mutation only causes a 4% chance of increased risk for a disease. Then off the wall mutations might pop up that say you carry a mutation where everyone dies by age 3. Well that one is clearly wrong. Then keep in mind that 23andme and even medical labs have been found to be wrong in the mutations they report people as having. No hyperventilating over anything, until everything has been fully vetted and checked and double checked and lab results confirmed by a medical lab.
Most things will be noted as interesting, yet, put in the back of your mind as something to be remembered.
If you click on the arrow next to "Pathogenic" or "Risk Factor" up will pop up the trait name. This one above comes up Becker Muscular Dystrophy. So then of course, off I go Googling "Becker Muscular Dystrophy" and it states --
"Becker muscular dystrophy is an X-linked recessive inherited disorder characterized by slowly progressive muscle weakness of the legs and pelvis. It is a type of dystrophinopathy. This is caused by mutations in the dystrophin gene, which encodes the protein dystrophin."
Well that's interesting. It falls under Mitochondrial Disease and fits some family symptoms. Definitely something I'd want to bring up with the Mito Doctor.
So I do that for each one. And it's time consuming and something I have to keep coming back to over time. I put the printout of the list in a folder and then for each one that pops up, I put why I discarded it or why it's interesting.
Please note above, that one of them is listed as "SNP x2". That's important. Only 5.17% of the population has one of those mutations and I have two of those mutations. Being homozygous (two of them) for a rare mutation can be more concerning for disease causing mutations. Spend extra time ruling those one's out. The mutation above with x2 is for an increased risk of inflammatory bowel disease.
Then for all the interesting mutations that I noted, I then look to see if they are on any genes noted by the doctors to cause Mitochondrial Disease. For example, for the Becker's Muscular Dystrophy mutation, I'm looking for the gene MT-ND1. For the Inflammatory Bowel Disease, I'm looking for NOD2. So I go to:
https://mseqdr.org/ and I click around until I get to the genes list (or I go directly by clicking on)
https://mseqdr.org/mb.php?url=index.php
I enter MT-ND1 in the spot for "Symbol" and I push enter. Up pops this.
What this says is that Mutations on the MT-ND1 gene have been reported by Mitochondrial Disease experts as causing Mitochondrial Disease. Under the columns for variants, it states that experts have entered 87 variants -- so a lot of Mito has been found due to variants on this gene. The column for Associated Diseases include LHON, MELAS.....
So then click on the "87" and up pops this page.
Where it says "Total Number of Public Variances Reported" click on the number. In this case it's click on 87. Then up pops the 87 variants that are currently entered into the database. You'll need to do it yourself to see all 87.
Next I want to see if my MT-ND1 mutation has been entered in the database. If it's not, it might mean they just haven't come across it yet, or it's not entered yet, or it's on a list at some university still waiting to be studied.
To find the DNA change name I have for my mutation, I have to go back to that Enlis page (above), where it has Becker's Muscular Dystrophy. At the beginning of that line it says it's Chromosome Position "M: 3,394". So I click on M: 3,394 on that Enlis page and up pops the name.
Continued.....