How PCR was first calibrated to find XMRV by Urisman et al [1]
First off, the Virochip technique was used to screen RNA samples isolated from prostate tumours. Notice that XMRV was not isolated by PCR and there was no attempt to target viral DNA and we are not talking about isolating XMRV from blood.
The RNA was amplified and purified into something called polyadenylated RNA. This is free of any DNA or anything else like histones. It is viral RNA (if present) and human RNA. This was then converted into cDNA and the fragments were isolated using something called a microarray.
This next bit is a bit technical but its meaning is crucial: Briefly, amplified nucleic acid from the tumour tissue, which hybridized to viral microarray oligonucleotides, was eluted from two specific spots.
The eluted DNA was re-amplified, and plasmid libraries constructed from this material were screened by colony hybridization using the spots' oligonucleotides as probes. The array oligonucleotides used in this case derived from the LTR region of murine type C retrovirus (MTCR) and spleen focus-forming virus (SFFV) [2].
The largest recovered fragment was 415 nt in length, and had 96% nt identity to the LTR region of MTCR, notice the multiple rounds of amplification. This is not PCR and the viral nucleic acid was picked up by primers complimentary for a LTR region.
Now most people realise that the XMRV genome is made up of GAG POL and ENV. This however is not the whole story. The genome has two identical regions of RNA at each side of the GAG POL ENV sequences. Now about 8% of our DNA is made up of endogenous retroviral sequences.
The reason that we are not all dead is because these babies cant replicate. Some of these are classed as MLV endogenous retroviruses. The key thing to remember that these ERVs have deletions and mutations which mean that they can't make the proteins needed to make virus particles. Now bearing this in mind I present the following diagram
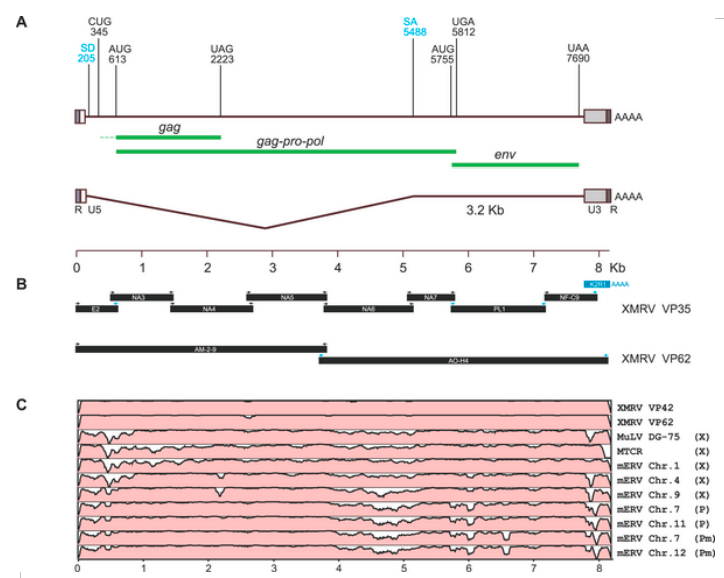
Now the upside down mountains are deletions and hence the HERVs can't code for proteins of the length needed. Bearing in mind that the original samples only contained RNA where would a non human ERV have come from? As XMRV can code for the full compliment of proteins needed to construct a virion it cannot by definition be a ERV. It is amazing how the Coffin cabal have missed this point.
Naked viral RNA cannot survive in a cellular environment and hence cannot be a contaminant. If there was contaminant DNA then the detection method used would not have selected it and it would not have entered the experimental environment.
As a final point in this instalment the authors in this study developed a PCR assay which could detect viral sequences in patient tissue known to be infected using the original micro array technique. They were able to do this by constructing primers complimentary to the gag sequences isolated by the other method.
They did this by adjusting the cycling conditions until they could detect the virus in DNA made from the RNA they isolated from the patients tissue. They had the advantage of knowing that the virus was present, and could change the parameter of the PCR reaction until they found it. They also used a form of PCR called reverse transcription PCR. That is for another time. Note the presence of mouse DNA is an impossibility as the process began with polyadenylated RNA!
1] Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, et al: Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog 2006, 2:e25.
2] Clark SP, Mak TW (1983) Complete nucleotide sequence of an infectious clone of Friend spleen focus-forming provirus: gp55 is an envelope fusion glycoprotein. Proc Natl Acad Sci U S A 80: 5037–5041.
First off, the Virochip technique was used to screen RNA samples isolated from prostate tumours. Notice that XMRV was not isolated by PCR and there was no attempt to target viral DNA and we are not talking about isolating XMRV from blood.
The RNA was amplified and purified into something called polyadenylated RNA. This is free of any DNA or anything else like histones. It is viral RNA (if present) and human RNA. This was then converted into cDNA and the fragments were isolated using something called a microarray.
This next bit is a bit technical but its meaning is crucial: Briefly, amplified nucleic acid from the tumour tissue, which hybridized to viral microarray oligonucleotides, was eluted from two specific spots.
The eluted DNA was re-amplified, and plasmid libraries constructed from this material were screened by colony hybridization using the spots' oligonucleotides as probes. The array oligonucleotides used in this case derived from the LTR region of murine type C retrovirus (MTCR) and spleen focus-forming virus (SFFV) [2].
The largest recovered fragment was 415 nt in length, and had 96% nt identity to the LTR region of MTCR, notice the multiple rounds of amplification. This is not PCR and the viral nucleic acid was picked up by primers complimentary for a LTR region.
Now most people realise that the XMRV genome is made up of GAG POL and ENV. This however is not the whole story. The genome has two identical regions of RNA at each side of the GAG POL ENV sequences. Now about 8% of our DNA is made up of endogenous retroviral sequences.
The reason that we are not all dead is because these babies cant replicate. Some of these are classed as MLV endogenous retroviruses. The key thing to remember that these ERVs have deletions and mutations which mean that they can't make the proteins needed to make virus particles. Now bearing this in mind I present the following diagram

Now the upside down mountains are deletions and hence the HERVs can't code for proteins of the length needed. Bearing in mind that the original samples only contained RNA where would a non human ERV have come from? As XMRV can code for the full compliment of proteins needed to construct a virion it cannot by definition be a ERV. It is amazing how the Coffin cabal have missed this point.
Naked viral RNA cannot survive in a cellular environment and hence cannot be a contaminant. If there was contaminant DNA then the detection method used would not have selected it and it would not have entered the experimental environment.
As a final point in this instalment the authors in this study developed a PCR assay which could detect viral sequences in patient tissue known to be infected using the original micro array technique. They were able to do this by constructing primers complimentary to the gag sequences isolated by the other method.
They did this by adjusting the cycling conditions until they could detect the virus in DNA made from the RNA they isolated from the patients tissue. They had the advantage of knowing that the virus was present, and could change the parameter of the PCR reaction until they found it. They also used a form of PCR called reverse transcription PCR. That is for another time. Note the presence of mouse DNA is an impossibility as the process began with polyadenylated RNA!
1] Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, et al: Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog 2006, 2:e25.
2] Clark SP, Mak TW (1983) Complete nucleotide sequence of an infectious clone of Friend spleen focus-forming provirus: gp55 is an envelope fusion glycoprotein. Proc Natl Acad Sci U S A 80: 5037–5041.
