Simon
Senior Member
- Messages
- 3,789
- Location
- Monmouth, UK
Guest blog by Professor Chris Ponting and colleagues at ME/CFS Research Review
UK Biobank - a national biobank different from the ME/CFS biobank - has data from around 500,000 individuals, including both healthy people and those with one or more of the many different diseases in the UK population. About 2,000 people in the sample reported that they had been given a diagnosis of CFS.
Analysis of data from this biobank indicates an inherited biological component for ME/CFS. The results show only one statistically significant change in a particular section of DNA and even this is problematic. This analysis indicates that a much bigger study, with many more ME/CFS cases, will be needed to indicate which genes and biological pathways are altered in people with ME/CFS.
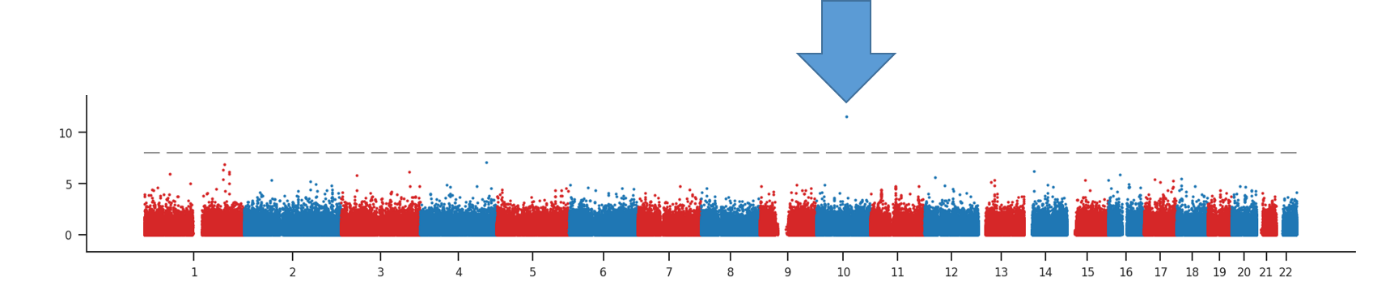
Statistical significance for the association between each DNA position and ME/CFS across 22 chromosomes. The arrow highlights the one “significant hit”.
Introduction
Myalgic encephalomyelitis (ME, also described as chronic fatigue syndrome, CFS) is a devastating long-term condition affecting 250,000 UK individuals. People with ME experience severe, disabling fatigue associated with post-exertional malaise. A few make good progress and may recover, while most others remain ill for years and may never recover. There is no known cause, or effective treatment for most. Consequently, it is vital to try new approaches to understand the reasons for the development of the condition.
This blog sets out what we can glean from the release, last summer, of data from about 500,000 individuals who make up the UK Biobank. (This biobank is not to be confused with the UK ME/CFS Biobank, UKMEB.) The data were acquired from individuals between 40 and 69 years of age in 2006-2010 who live across the UK. These people provided samples (e.g. blood, urine and saliva) and answered questionnaires. In addition, for some of these people their electronic health record data are being linked in. Importantly for this blog, the DNA variation (‘genotype’) of all the volunteer participants has been determined.
Genetic variation can provide insights into the causes of disease when these have a heritable component (i.e. are inherited down through the generations). DNA sequence is not altered by disease (except in cancer) and so variants can reveal the causes, rather than consequences, of disease.
....
Read the full blog
UK Biobank - a national biobank different from the ME/CFS biobank - has data from around 500,000 individuals, including both healthy people and those with one or more of the many different diseases in the UK population. About 2,000 people in the sample reported that they had been given a diagnosis of CFS.
Analysis of data from this biobank indicates an inherited biological component for ME/CFS. The results show only one statistically significant change in a particular section of DNA and even this is problematic. This analysis indicates that a much bigger study, with many more ME/CFS cases, will be needed to indicate which genes and biological pathways are altered in people with ME/CFS.

Statistical significance for the association between each DNA position and ME/CFS across 22 chromosomes. The arrow highlights the one “significant hit”.
Introduction
Myalgic encephalomyelitis (ME, also described as chronic fatigue syndrome, CFS) is a devastating long-term condition affecting 250,000 UK individuals. People with ME experience severe, disabling fatigue associated with post-exertional malaise. A few make good progress and may recover, while most others remain ill for years and may never recover. There is no known cause, or effective treatment for most. Consequently, it is vital to try new approaches to understand the reasons for the development of the condition.
This blog sets out what we can glean from the release, last summer, of data from about 500,000 individuals who make up the UK Biobank. (This biobank is not to be confused with the UK ME/CFS Biobank, UKMEB.) The data were acquired from individuals between 40 and 69 years of age in 2006-2010 who live across the UK. These people provided samples (e.g. blood, urine and saliva) and answered questionnaires. In addition, for some of these people their electronic health record data are being linked in. Importantly for this blog, the DNA variation (‘genotype’) of all the volunteer participants has been determined.
Genetic variation can provide insights into the causes of disease when these have a heritable component (i.e. are inherited down through the generations). DNA sequence is not altered by disease (except in cancer) and so variants can reveal the causes, rather than consequences, of disease.
....
Read the full blog
