JaimeS
Senior Member
- Messages
- 3,408
- Location
- Silicon Valley, CA
"The scientists also discovered that people with C.F.S. had higher blood levels of lipopolysaccharides, inflammatory molecules that may indicate that bacteria have moved from the gut into the bloodstream, where they can produce various symptoms of disease."
are lipopolysaccharides in the bloodstream proof of leaky gut?
Elevated LPS from bacteria you'd expect to see in the gut would be. I think they're just not calling it 'leaky gut' because the alt. medicine practitioners pioneered the idea of inflammation causing leaky gut and leading to food intolerances (the latter of which is still 'imaginary' in the minds of many mainstream practitioners). Now there is better evidence that this is a common phenomena, but mainstream med has to call it something else, or it will keep the alt med connection in people's minds. It's a 're-branding' of an old idea.
Also, the researchers had other markers that specifically indicated gut damage, apart from LPS.
What are 'firmicutes'? Are they the Beneficial Bacteria in the gut?
Firmicutes is a huge and inspecific category that contains many kinds of bacteria, some of which are associated with illness, some of which are associated with good health.
My Genova test did not mention firmicutes.
Perhaps they broke things down more or less specifically. Clostridium, e.g. are Firmucutes -- bet they mentioned those.
I had RedLabs stool test done last year. My bacterial findings were identical to those in the study -- even the inflammatory markers followed the patterns they talk about. The only difference is that my level of bacterial diversity was exactly mid-range normal. I had almost never in my life taken antibiotics at that time. After long-term abx, I wonder what the story would be, now.
My article on this study and other gut studies is up on #MEAction as of yesterday -- it has more info than I've put here.
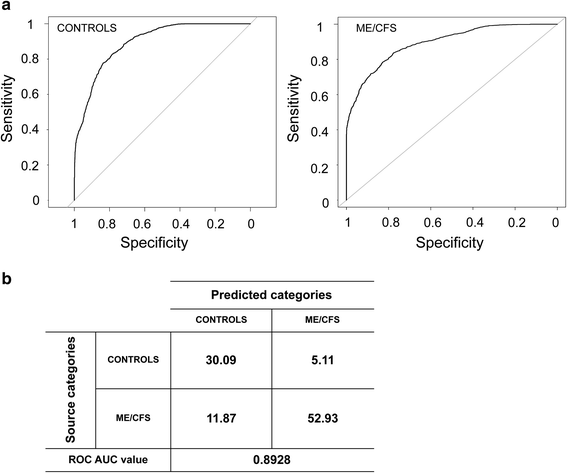
New research: gut microbes identify 83% of patients

July 13, 2016
- By Jaime S
Last edited: