RogerBlack
Senior Member
- Messages
- 902
http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1006292
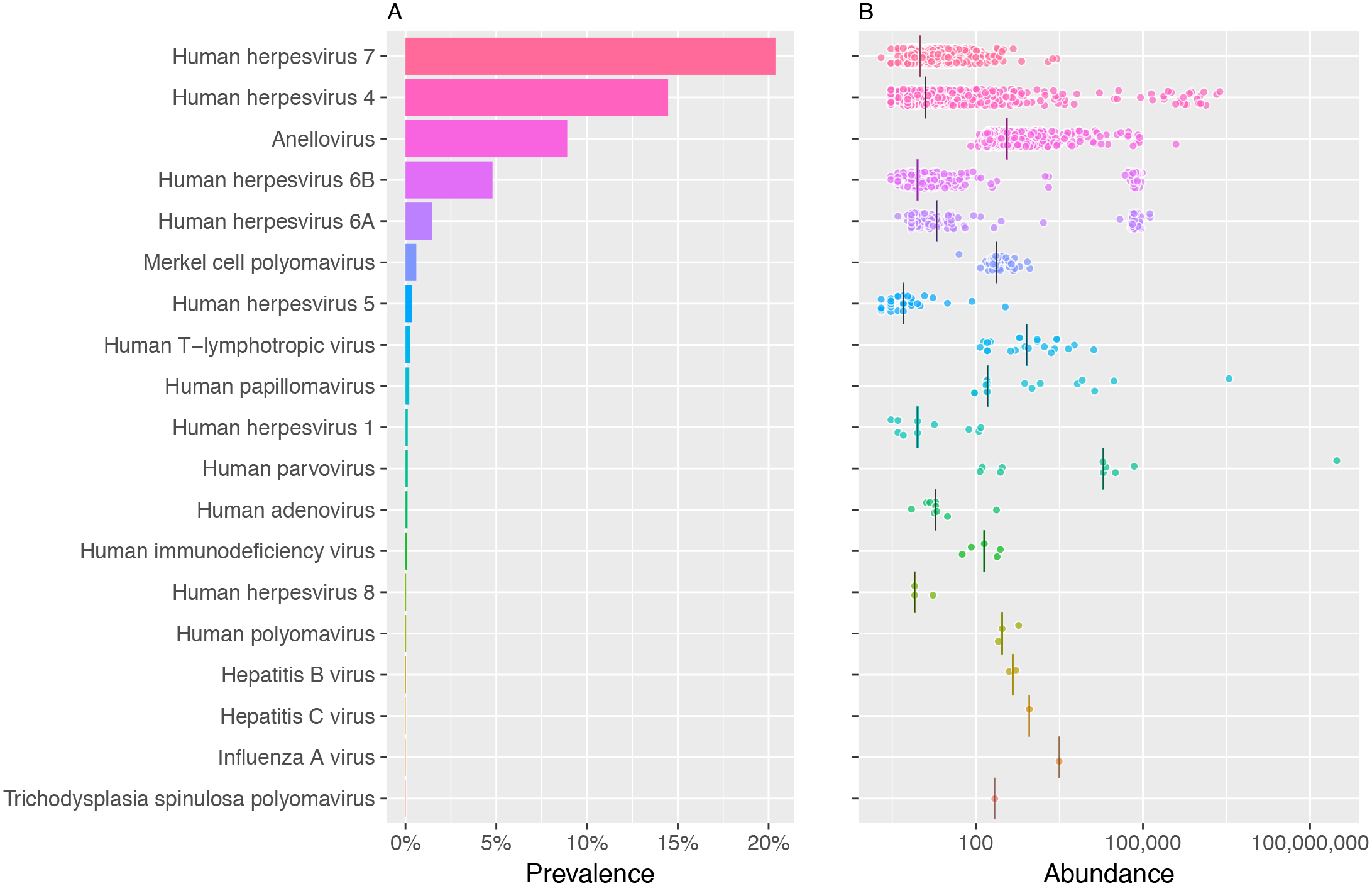
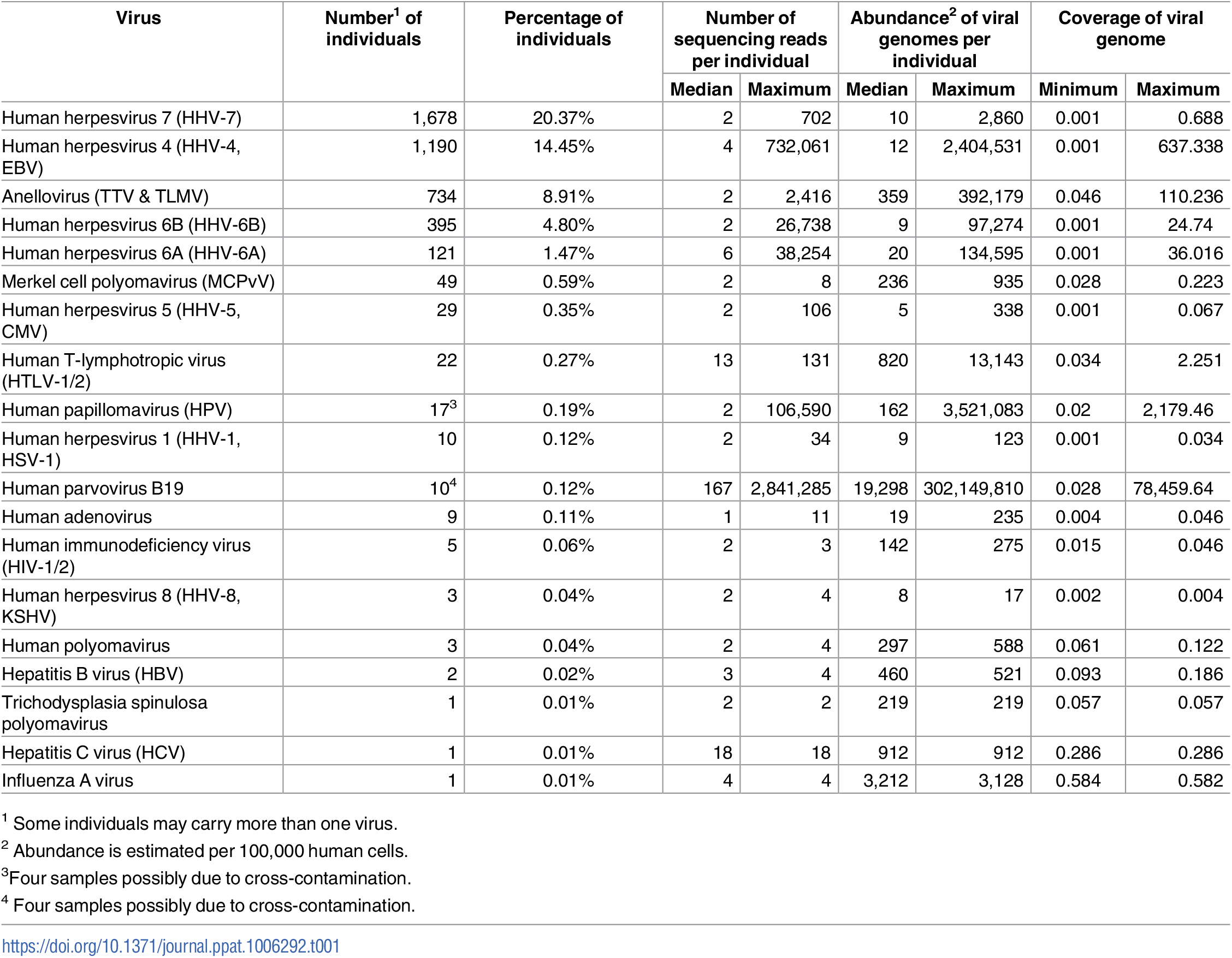
A graph of how many people in the sample each virus is found in, and the estimate of abundance per 100000 human blood cells.
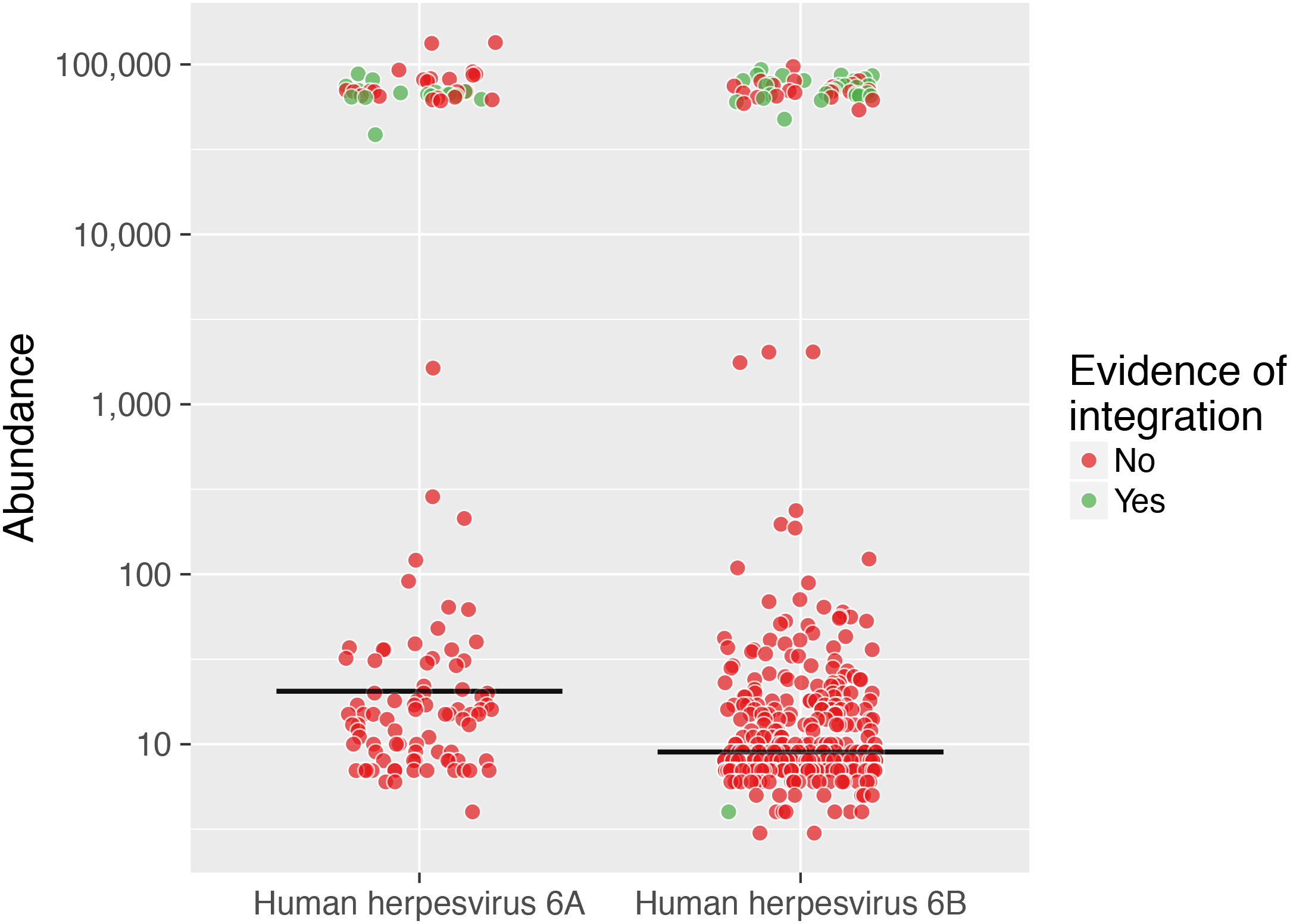
Some viruses can be in a latent form, integrated into human DNA, to remain latent for long periods. This shows how many are integrated versus abundance.
Caveats: This study was not aimed at finding either rare, or the similar population of RNA viruses that exist. It also shows the problem with contamination, in that most of the viruses found (70) were lab containments, with some from well understood and expected sources used for calibration, but others from a wide variety of lab sources.
Here, we identify the viral DNA sequences in blood of over 8,000 individuals undergoing whole genome sequencing. This approach serves to identify 94 viruses; however, many are shown to reflect widespread DNA contamination of commercial reagents or of environmental origin. While this represents a significant limitation to reliably identify novel viruses infecting humans, we could confidently detect sequences and quantify abundance of 19 human viruses in 42% of individuals. Ancestry, sex, and age were important determinants of viral prevalence. This large study calls attention on the challenge of interpreting next generation sequencing data for the identification of novel viruses. However, it serves to categorize the abundance of human DNA viruses using an unbiased technique.
A graph of how many people in the sample each virus is found in, and the estimate of abundance per 100000 human blood cells.
Some viruses can be in a latent form, integrated into human DNA, to remain latent for long periods. This shows how many are integrated versus abundance.
Caveats: This study was not aimed at finding either rare, or the similar population of RNA viruses that exist. It also shows the problem with contamination, in that most of the viruses found (70) were lab containments, with some from well understood and expected sources used for calibration, but others from a wide variety of lab sources.
