First of all, huge thanks to @Valentijn , without whom this post - and likely a majority of the ones before it - would not exist. You can find the majority of the information here in the comments section of the previous post.
So, how do you tell if the protein structure has been altered in a meaningful way, or at all, if there is literally no clinical research on your SNP? Let's be honest - we would rarely be interested in going so far. However, looking at last week's missense mutation on Von Willebrand Clotting Factor - well, it'd be a good idea to know how seriously to take that SNP.
Using BLOSUM62:
The first and quickest way to determine the impact of a missense mutation is to use a BLOSUM62 chart. The chart is based off of how dire it is to exchange one amino acid for another. If the amino acids are similarly-shaped (and behave similarly enough) even a missense mutation might not cause a major problem. Last time, we noted that my VWF SNP exchanged proline for serine, so we use the chart to find where proline and serine cross... The higher the score, the more likely the amino acids will behave similarly enough that there isn't a problem. The lower the value on the chart, the more the SNP is a potential concern.
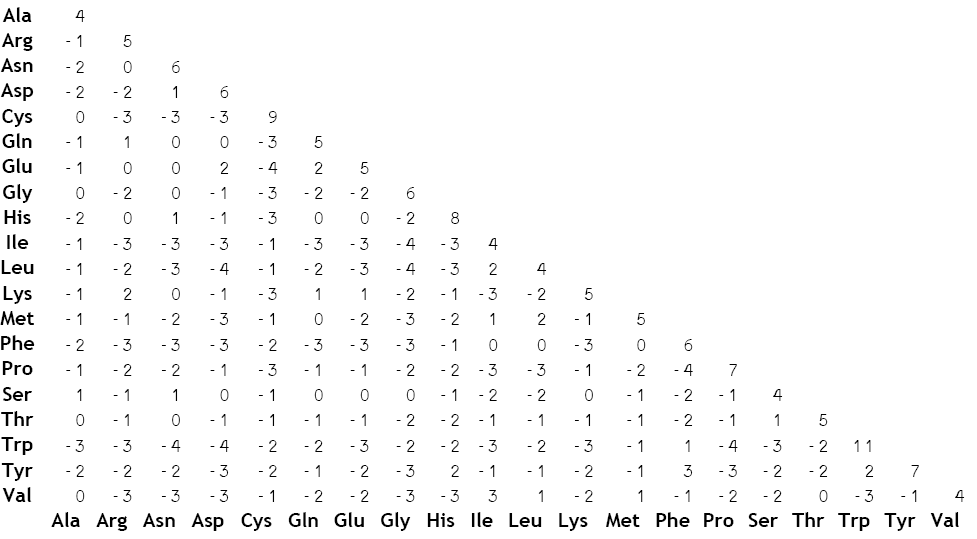
(Hannes Rost, 2008)
Finding where proline and serine meet, we see that this has a value of -1. Here are the possible values from the chart:
This is not a negative 4, 3, or 2, so it's not dire - but it's not marvelous, either. This is the kind of inconclusive thing that is bothersome enough that we can take it to the final and most complicated step to discover if this mutation is really going to cause clotting issues...
Using a Three-Dimensional Modelling Program:
Let's start with a quote from Part II of this series:
What we're really looking for is the answer to the question: does this amino acid substitution significantly change the shape of the protein - enough to affect its function? Or perhaps the shift is minor but in a very important location, like at a receptor site?
Three-dimensional modelling can answer that question!
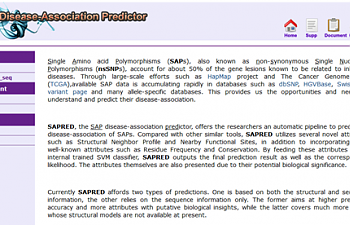
The SAP Disease-Association Predictor
I used the SAP Disease-Association Predictor. You will have to get an account with them using a valid email address, but the account is otherwise free, and so far they haven't sent me a single advertisement for anything. After you've sent them an email and they've replied back with a link to activate your account, you will find yourself here:
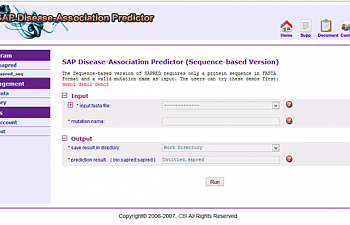
This is the introductory page. Read about what an SNP is and all kind of other introductory information! Or click right where it says 'Run sapred_seq' on the left-hand side.
Next, you want to be able to '* input fasta file'. But we don't yet have a FASTA file. We can, however, get one.
Generating the FASTA File:
Not actually tough, I promise. First, you're going to go to www.uniprot.org to search for the gene, but keep the previous window open. You'll need the gene's proper abbreviation, which is typically three letters. In the case of Von Willebrand Factor, it's VWF. Rather unsurprisingly. Put that into the search box and click 'search'.
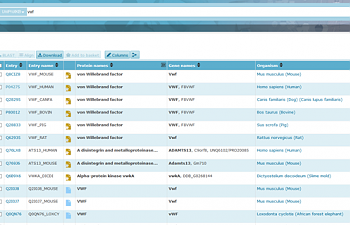
You'll notice that you get a bunch of links rather than just one. That's because uniprot lists not just humans, but a variety of organisms that have a Von Willebrand Factor. Make sure that you click on the one that says 'human'!
You should see the following screen if you clicked on the proper thing:
If this isn't what you see, you might have clicked on the mouse one, or the actual thing that says Homo sapiens, which will take you to stuff about people in general. You should have clicked on the link that says 'P04275'.
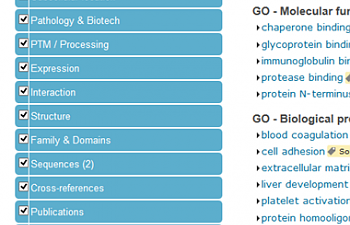
This actually is pretty interesting; it says that VWF is actually involved in a variety of molecular and biological processes and you can learn a lot about them. I'd save that for if the VWF SNP is significant. If it's not, we're wasting precious research time.
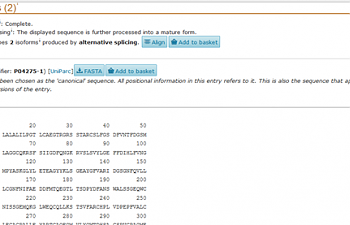
So: see the bright blue area to the left that reads 'Display'? Find 'Sequences' and click on it.
Yes! Now we should be here:
Click where it says 'FASTA'.
A bunch of alphanumerics should pop up. Believe it or not, this actually represents the amino acids in the correct order in the VWF protein. This is what we want. Select everything on the screen and copy it.
Go back to your (hopefully still open) SAP Disease Predictor. Click on input fasta and paste your code into the area where it says 'OR paste into window'.
Naming the Mutation:
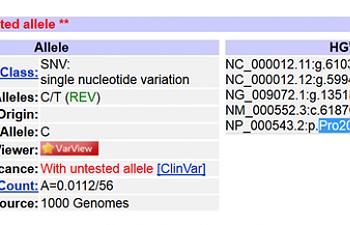
Finally, we need the name of the mutation. It will include the first letter of the 'correct' amino acid and the one that's taken its place in this SNP. It's locate-able through dbSNP most rapidly. Use dbSNP to search for rs61750615 and click on the link for rs61750615. Once again, you'll see that the mutation is Pro2063Ser. Ditch all but the first letter of the amino acid and this becomes P2063S.
This is the name of the mutation, so far as SAP Disease Predictor is concerned. For some amino acids it won't quite work this way; for example, both leucine and lysine can't be represented with 'L'. Go here to see how to write the correct one-letter symbol for the amino acid you need.
Generating the Report:
If you're planning on doing more than one of these, click where it says 'Untitled' and change the file name to something meaningful like 'Von Willebrand SNP'.
Input the name of the mutation where it says to do so.
Click 'run'.
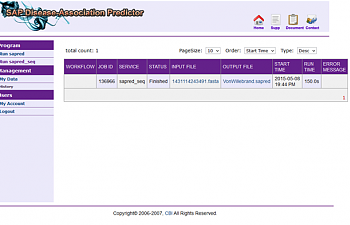
So now we wait. VWF is actually a pretty large protein, so the analysis is going to take a few minutes. The program will send you an email once the analysis is complete, so do something that'll take about five minutes.
Yes! Complete!
...to be continued
<-------Previous Entry
<---Start of the Series
So, how do you tell if the protein structure has been altered in a meaningful way, or at all, if there is literally no clinical research on your SNP? Let's be honest - we would rarely be interested in going so far. However, looking at last week's missense mutation on Von Willebrand Clotting Factor - well, it'd be a good idea to know how seriously to take that SNP.
Using BLOSUM62:
The first and quickest way to determine the impact of a missense mutation is to use a BLOSUM62 chart. The chart is based off of how dire it is to exchange one amino acid for another. If the amino acids are similarly-shaped (and behave similarly enough) even a missense mutation might not cause a major problem. Last time, we noted that my VWF SNP exchanged proline for serine, so we use the chart to find where proline and serine cross... The higher the score, the more likely the amino acids will behave similarly enough that there isn't a problem. The lower the value on the chart, the more the SNP is a potential concern.
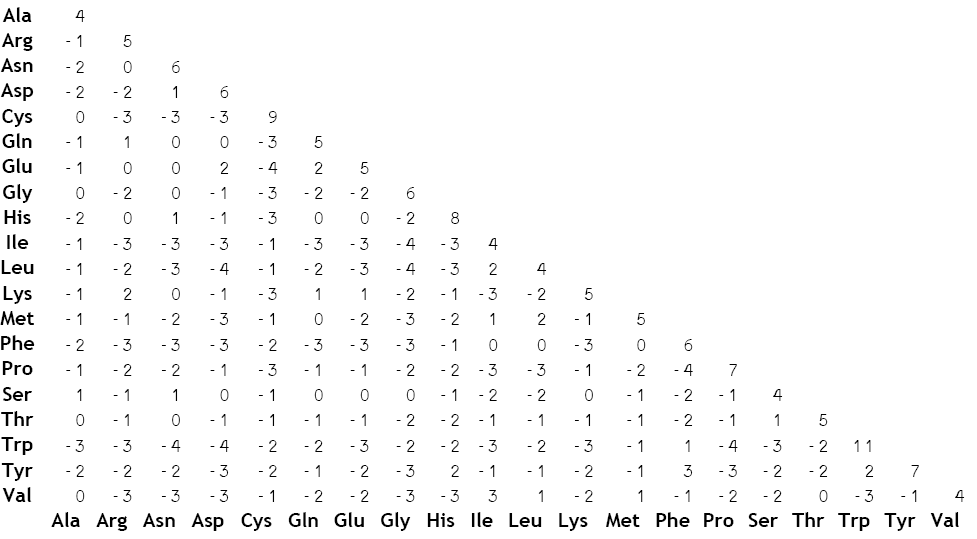
(Hannes Rost, 2008)
Finding where proline and serine meet, we see that this has a value of -1. Here are the possible values from the chart:
-4 -3 -2 -1 0 1 2 3 4
This is not a negative 4, 3, or 2, so it's not dire - but it's not marvelous, either. This is the kind of inconclusive thing that is bothersome enough that we can take it to the final and most complicated step to discover if this mutation is really going to cause clotting issues...
Using a Three-Dimensional Modelling Program:
Let's start with a quote from Part II of this series:
What we're really looking for is the answer to the question: does this amino acid substitution significantly change the shape of the protein - enough to affect its function? Or perhaps the shift is minor but in a very important location, like at a receptor site?
Three-dimensional modelling can answer that question!
The SAP Disease-Association Predictor
I used the SAP Disease-Association Predictor. You will have to get an account with them using a valid email address, but the account is otherwise free, and so far they haven't sent me a single advertisement for anything. After you've sent them an email and they've replied back with a link to activate your account, you will find yourself here:
This is the introductory page. Read about what an SNP is and all kind of other introductory information! Or click right where it says 'Run sapred_seq' on the left-hand side.
Next, you want to be able to '* input fasta file'. But we don't yet have a FASTA file. We can, however, get one.
Generating the FASTA File:
Not actually tough, I promise. First, you're going to go to www.uniprot.org to search for the gene, but keep the previous window open. You'll need the gene's proper abbreviation, which is typically three letters. In the case of Von Willebrand Factor, it's VWF. Rather unsurprisingly. Put that into the search box and click 'search'.
You'll notice that you get a bunch of links rather than just one. That's because uniprot lists not just humans, but a variety of organisms that have a Von Willebrand Factor. Make sure that you click on the one that says 'human'!
You should see the following screen if you clicked on the proper thing:
If this isn't what you see, you might have clicked on the mouse one, or the actual thing that says Homo sapiens, which will take you to stuff about people in general. You should have clicked on the link that says 'P04275'.
This actually is pretty interesting; it says that VWF is actually involved in a variety of molecular and biological processes and you can learn a lot about them. I'd save that for if the VWF SNP is significant. If it's not, we're wasting precious research time.
So: see the bright blue area to the left that reads 'Display'? Find 'Sequences' and click on it.
Yes! Now we should be here:
Click where it says 'FASTA'.
A bunch of alphanumerics should pop up. Believe it or not, this actually represents the amino acids in the correct order in the VWF protein. This is what we want. Select everything on the screen and copy it.
Go back to your (hopefully still open) SAP Disease Predictor. Click on input fasta and paste your code into the area where it says 'OR paste into window'.
Naming the Mutation:
Finally, we need the name of the mutation. It will include the first letter of the 'correct' amino acid and the one that's taken its place in this SNP. It's locate-able through dbSNP most rapidly. Use dbSNP to search for rs61750615 and click on the link for rs61750615. Once again, you'll see that the mutation is Pro2063Ser. Ditch all but the first letter of the amino acid and this becomes P2063S.
This is the name of the mutation, so far as SAP Disease Predictor is concerned. For some amino acids it won't quite work this way; for example, both leucine and lysine can't be represented with 'L'. Go here to see how to write the correct one-letter symbol for the amino acid you need.
Generating the Report:
If you're planning on doing more than one of these, click where it says 'Untitled' and change the file name to something meaningful like 'Von Willebrand SNP'.
Input the name of the mutation where it says to do so.
Click 'run'.
So now we wait. VWF is actually a pretty large protein, so the analysis is going to take a few minutes. The program will send you an email once the analysis is complete, so do something that'll take about five minutes.
Yes! Complete!
...to be continued
<-------Previous Entry
<---Start of the Series